Minor Version: 8.4.4 (May 1, 2018) Introduction The Tinker molecular modeling software is a complete and general package for molecular mechanics and dynamics, with some special features for biopolymers. Tinker has the ability to use any of several common parameter sets, such as Amber (ff94, ff96, ff98, ff99, ff99SB), CHARMM (19, 22, 22/CMAP). 63 rows Comparison of software for molecular mechanics modeling Jump to navigation Jump to.
AMBER Molecular Dynamics
Contents
- AMBER Roll
This roll installs AMBER tools.There are 4 flavors compiled for each compiler (gnu, intel):
Amber is a licensed software. Get licensed files for AMBAER and AmberTools fromAmarolab and save copy in nbcr.ucsd@gmail.com accountDownload the appropriate amber source files into the src/amber.Download the benchmark suite in src/amber-test:
Dependencies
- The
mpiroll must be installed on the build/install host.depends on mpi libraries. - The
intelroll must be installed on the build/install host. In the absence of intel compilerthe roll should be buld withROLLCOMPILEr='gnu'. - The
cudaroll must be installed on build/install host . In the absence of cuda, editsrc/amber/Makefiletargets to remove cuda-specific compilation. - The roll sources assumes that modulefiles provided by
intel,cudaandmpirolls are available.
The roll uses openmpi compiled with intel and gnu compilers for eth fabric.To make a roll use the following command:
A successful build will create the file amber-*.disk1.iso.
To install, execute these instructions on a Rocks frontend:
Tthe roll installs amber and environment module files in:
For Sep 2015 workshop buid amberGaMD roll based on a received patch for amber14 sourceCreate the following changes to the roll source:
See diffs for the above files in GaMD-diffs file at the top of the roll.After creating changes build amberGaMD roll
Testing
A test script amber.t can be run to verify properinstallation of the roll binaries and module files. Torun the test scripts execute the following command(s):
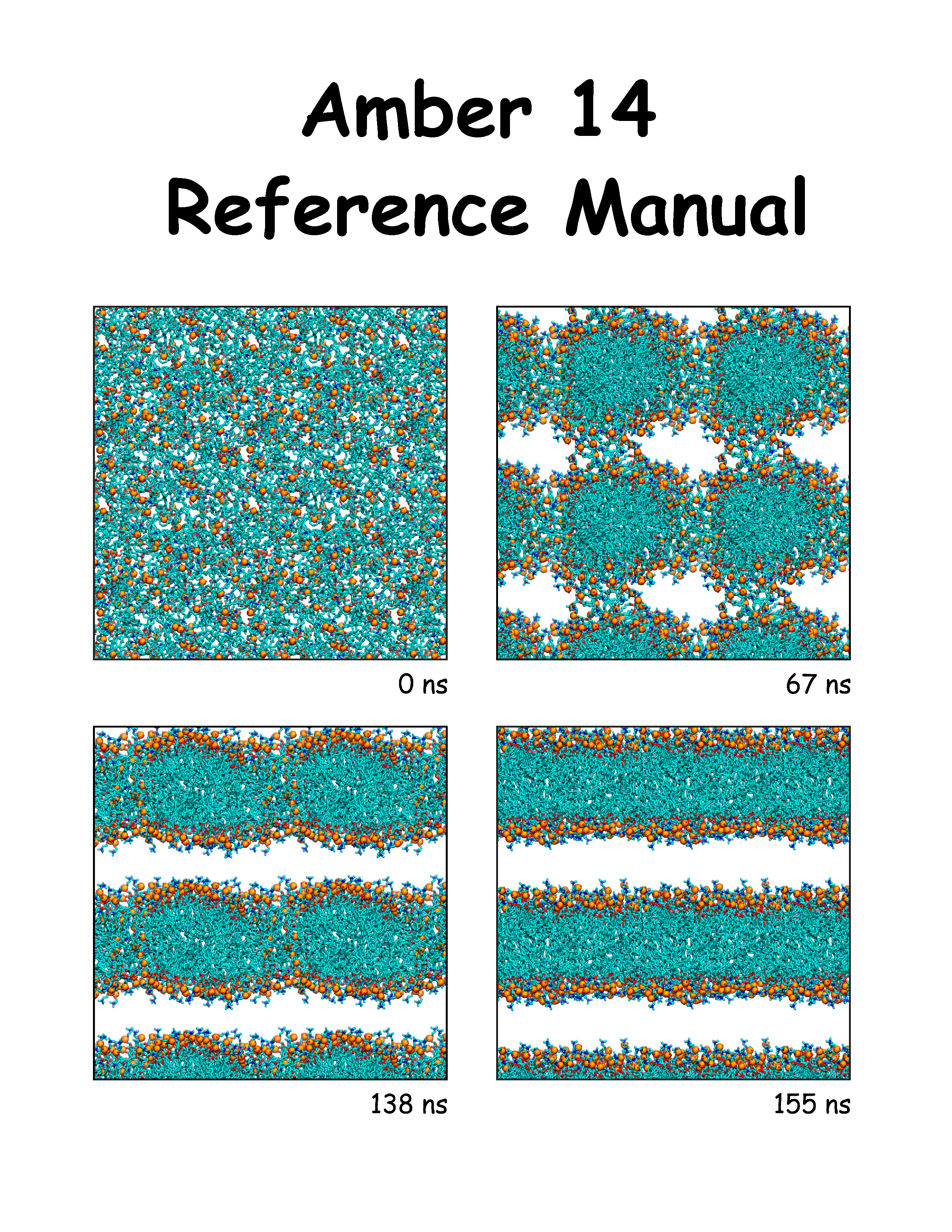
A benchmark suit distro (from Amber site) contains a shell script run_bench_CPU+GPU.shand required input to run suite of tests. Untar, adjust cpu and gpu counts in the scriptfor a specific host.
This is a list of computer programs that are predominantly used for molecular mechanics calculations.
- GPU – GPU accelerated
- I – Has interface
- Imp – Implicit water
- MC – Monte Carlo
- MD – Molecular dynamics
- Min – Optimization
- QM – Quantum mechanics
- REM – Replica exchange method
| Name | View 3D | Model builder | Min | MD | MC | REM | QM | Imp | GPU | Comments | License | Website |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abalone | Yes | Yes | Yes | Yes | Yes | Yes | I | Yes | Yes | Biomolecular simulations, protein folding. | Proprietary, gratis, commercial | Agile Molecule |
| ACEMD[1] | No | No | Yes | Yes | No | Yes | No | No | Yes | Molecular dynamics with CHARMM, Amber forcefields; runs on NVIDIA GPUs, highly optimized with CUDA | Proprietary, gratis and commercial versions | Acellera Ltd[permanent dead link] |
| ADF | Yes | Yes | Yes | Yes | No | No | Yes | Yes | Yes | Modeling suite: ReaxFF, UFF, QM-MM with Amber and Tripos force fields, DFT and semi-empirical methods, conformational analysis with RDKit; partly GPU-accelerated | Proprietary, commercial, gratis trial | SCM |
| ADUN[2] | No | No | Yes | Yes | No | No | Yes | Yes | Yes | Charmm, AMBER, user specified (via force field markup language, FFML), QM-MM calculations with empirical valence bond (EVB); framework based (GNUStep-Cocoa); SCAAS for spherical boundary conditions | Free open sourceGNU GPL | https://web.archive.org/web/20160303133843/https://gna.org/projects/adun |
| Assisted Model Building with Energy Refinement (AMBER)[3] | No | Yes | Yes | Yes | No | Yes | I | Yes | Yes | ? | Proprietary | ambermd.org |
| Ascalaph Designer | Yes | Yes | Yes | Yes | Yes | Yes | I | Yes | Yes | Molecular building (DNA, proteins, hydrocarbons, nanotubes), molecular dynamics, GPU acceleration | Mixed: free open source (GNU GPL) & commercial | Ascalaph Project |
| Automated Topology Builder (ATB) | Yes | Yes | Yes | No | No | No | No | No | No | Automated molecular topology building service for small molecules (< 99 atoms). GROMOS, GROMACS, CNS, LAMMPS formats with validation Repository for molecular topologies and pre-equilibrated systems | Proprietarycommercial, gratis academic use | Automated Topology Builder |
| Avogadro | Yes | Yes | Yes | No | No | No | I | No | No | Molecule building, editing (peptides, small molecules, crystals), conformational analysis, 2D/3D conversion; extensible interfaces to other tools | Free open sourceGNU GPL | Avogadro |
| BOSS | No | No | Yes | No | Yes | No | Yes | No | No | OPLS | Proprietary | Yale University |
| CHARMM | No | Yes | Yes | Yes | Yes | I | I | Yes | Yes | Commercial version with multiple graphical front ends is sold by Accelrys (as CHARMm) | Proprietary, commercial | charmm.org |
| ChemDoodle 3D | Yes | Yes | Yes | No | No | No | No | No | No | Intuitive building of molecules in real-time; production of high quality publication graphics; works on Windows, macOS and Linux; developed by iChemLabs | Proprietary, commercial, free trial | ChemDoodle 3D Website |
| CHEMKIN | No | No | No | No | No | No | No | No | No | Chemical reaction kinetics. | Proprietary | CHEMKIN |
| COSMOS | Yes | Yes | Yes | Yes | Yes | No | I | No | No | Hybrid QM-MM COSMOS-NMR force field with fast semi-empirical calculation of electrostatic and/or NMR properties; 3-D graphical molecule builder and viewer | Proprietary, commercial, free without GUI | COSMOS Software |
| CP2K | No | No | Yes | Yes | Yes | No | Yes | Yes | Yes | CP2K can perform atomistic and molecular simulations of solid state, liquid and biological systems. | Free open sourceGNU GPLv2 or later | CP2K |
| Culgi | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | No | Atomistic and coarse-grained simulations, mesoscale methods, COSMO-RS and UNIFAC, database integration, mapping techniques, AM1 and interface to MOPAC/NWChem | Proprietarycommercial, gratis academic education | Culgi BV |
| Desmond | Yes | Yes | Yes | Yes | No | Yes | No | No | Yes | High performance MD; has comprehensive GUI to build, visualize, and review results and calculation setup up and launch | Proprietary, commercial or gratis | |
| DFTB+ | No | No | Yes | No | No | No | No | No | No | A fast and efficient versatile quantum mechanical simulation software package. Using DFTB+ you can carry out quantum mechanical simulations similar to density functional theory but in an approximate way, typically gaining around two orders of magnitude in speed. | Free open sourceGNU LGPL | [1] |
| Discovery Studio | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | No | Comprehensive life science modeling and simulation suite of applications focused on optimizing drug discovery process: small molecule simulations, QM-MM, pharmacophore modeling, QSAR, protein-ligand docking, protein homology modeling, sequence analysis, protein-protein docking, antibody modeling, etc. | Proprietary, trial available | Dassault Systèmes BIOVIA (formerly Accelrys) |
| DL_POLY | No | Yes | Yes | Yes | No | No | I | No | No | DL_POLY is a general purpose classical molecular dynamics (MD) simulation software developed at Daresbury Laboratory by I.T. Todorov and W. Smith. It is part of the DL_Software suite of projects including DL_MESO (DPD), DL_FIELD (FF builder), ChemShell (QMMM environment), etc. | Proprietarycommercial, free source for academic use | DL_POLY |
| Energy Calculation and Dynamics (ENCAD) | No | No | No | Yes | No | No | No | No | No | Notably the first MD simulation software by Michael Levitt | Free open source | [2] |
| fold.it | Y / I | Yes | Yes | Yes | Yes | Yes | I | No | No | University of Washington and The Baker Labs; structure prediction, protein folding | Proprietary, commercial or gratis | fold.it download page |
| FoldX | I | Yes | Yes | No | No | No | No | No | No | Energy calculations, protein design | Proprietary, commercial or gratis | CRG |
| GOMC | No | No | No | No | Yes | Yes | No | No | Yes | GPU Optimized Monte Carlo. Atomistic simulation engine, capable of simulating complex systems. | Free opensource.GNU Affero General Public License. | GOMC website. GOMC Github |
| GPIUTMD | I | I | Yes | Yes | No | I | No | No | Yes | Performs general purpose particle dynamics simulations on a single workstation, exploiting NVIDIA CUDA GPUs | Proprietary | http://gpiutmd.iut.ac.ir |
| GROMACS | No | No | Yes | Yes | No[4] | Yes | I | Yes[5] | Yes | High performance MD | Free open sourceGNU GPL | gromacs.org |
| GROMOS | No | No | Yes | Yes | Yes | Yes | No | Yes | Yes | Intended for biomolecules | Proprietary, commercial | GROMOS website |
| GULP | No | No | Yes | Yes | No | No | No | No | No | Molecular dynamics and lattice optimization | Proprietary, free academic use | http://gulp.curtin.edu.au/gulp/ |
| HOOMD-blue | No | No | Yes | Yes | Yes | No | No | No | Yes | General-purpose molecular dynamics highly optimized for GPUs, includes various pair potentials, Brownian dynamics, dissipative particle dynamics, rigid body constraints, energy minimizing, etc. | Free open source | http://glotzerlab.engin.umich.edu/hoomd-blue/ |
| HyperChem | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | No | De Facto Standard Molecular Modeling System | Proprietary | Hypercube, Inc. |
| ICM | Yes | Yes | Yes | No | Yes | No | I | Yes | No | Powerful global optimizer in an arbitrary subset of internal variables, NOEs, docking (protein, ligand, peptide), EM, density placement | Proprietary | Molsoft |
| LAMMPS | Yes | Yes | Yes | Yes | Yes | Yes | I | Yes | Yes | Has potentials for soft and solid-state materials and coarse-grain systems | Free open source, GNU GPLv2 | Sandia |
| MacroModel | Yes | Yes | Yes | Yes | Yes | No | I | Yes | No | OPLS-AA, MMFF, GBSA solvent model, conformational sampling, minimizing, MD. Includes the Maestro GUI which provides visualizing, molecule building, calculation setup, job launch and monitoring, project-level organizing of results, access to a suite of other modelling programs. | Proprietary | Schrödinger |
| MAPS[6] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Building, visualizing, and analysis tools in one user interface, with access to multiple simulation engines | Proprietary, trial available | Scienomics |
| Materials Studio | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Environment that brings materials simulation technology to desktop computing, solving key problems in R&D processes | Proprietary, trial available | Dassault Systèmes BIOVIA (formerly Accelrys) |
| MBN Explorer[7] + MBN Studio | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes | Standard and reactive CHARMM force fields; molecular modeler (carbon nanomaterials, biomolecules, nanocrystals); explicit library of examples | Proprietary, free trial available | MBN Research Center |
| MedeA | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No | Combines leading experimental databases and major computational programs like the Vienna Ab-initio Simulation Package (VASP), LAMMPS, GIBBS with sophisticated materials property prediction, analysis, visualizing | Proprietary, trial available | Materials Design |
| MCCCS Towhee | No | No | No | No | Yes | No | No | No | No | Originally designed to predict fluid phase equilibria | Free open sourceGNU GPL | Towhee Project |
| MDynaMix | No | No | No | Yes | No | No | No | No | No | Parallel MD | Free open sourceGNU GPL | Stockholm University |
| MOE | Yes | Yes | Yes | Yes | No | No | I | Yes | No | Molecular Operating Environment (MOE) | Proprietary | Chemical Computing Group |
| MOIL | Yes | Yes | Yes | Yes | No | Yes | No | No | Yes | Includes action-based algorithms (stochastic difference equations in time, length) and locally enhanced sampling | Free open source, public domain, source code | MOIL |
| MolMeccano | No | Yes | No | No | No | No | No | No | No | Semi-automatic Force Field parametrizer | Proprietary | MolMeccano Project |
| Orac | No | No | Yes | Yes | No | Yes | No | Yes | No | Molecular dynamics simulation program to explore free energy surfaces in biomolecular systems at the atomic level | Free open source | Orac download page |
| NAB[8] | No | Yes | No | No | No | No | No | No | No | Generates models for unusual DNA and RNA | Free open source | Case group |
| NAMD + VMD | Yes | Yes | Yes | Yes | No | Yes | I | Yes | Yes | Fast, parallel MD, CUDA | Proprietary, free academic use, source code | Beckman Institute |
| NWChem | No | No | Yes | Yes | No | No | Yes | No | No | High-performance computational chemistry software, includes quantum mechanics, molecular dynamics and combined QM-MM methods | Free open source, Educational Community License version 2.0 | NWChem |
| Packmol | No | Yes | No | No | No | No | No | No | No | Builds complex initial configurations for molecular dynamics | Free open sourceGNU | link |
| Prime | Yes | Yes | Yes | No | Yes | No | I | Yes | No | Homology modeling, loop and side chain optimizing, minimizing, OPLS-AA, SGB solvent model, parallalized | Proprietary | Schrödinger |
| Protein Local Optimization Program | No | Yes | Yes | Yes | Yes | No | No | No | No | Helix, loop, and side chain optimizing, fast energy minimizing | Proprietary | PLOP wiki |
| Q | No | No | No | Yes | No | No | No | No | No | (I) Free energy perturbation (FEP) simulations, (II) empirical valence bond (EVB), calculations of reaction free energies, (III) linear interaction energy (LIE) calculations of receptor-ligand binding affinities | Uppsala Molekylmekaniska HB | Q |
| RedMD[9] | I | Yes | Yes | Yes | Yes | No | No | No | No | Reduced MD, package for coarse-grained simulations | Free, open sourceGNU | University of Warsaw, ICM |
| SAMSON | Yes | Yes | Yes | Yes | No | No | Yes | No | No | Computational nanoscience (life sciences, materials, etc.). Modular architecture, modules termed SAMSON Elements | Proprietary, gratis | SAMSON Connect |
| StruMM3D (STR3DI32) | Yes | Yes | Yes | Yes | No | No | No | No | No | Sophisticated 3-D molecule builder and viewer, advanced structural analytical algorithms, full featured molecular modeling and quantitation of stereo-electronic effects, docking, and handling of complexes. | Proprietary, free 200 atom version | Exorga, Inc. |
| Scigress | Yes | Yes | Yes | Yes | No | No | Yes | Yes | No | MM, DFT, semiempirical methods, parallel MD, conformational analysis, Linear scaling SCF, docking protein-ligand, Batch processing, virtual screening, automated builders (molecular dynamics, proteins, crystals) | Proprietary | SCIGRESS.com |
| Spartan | Yes | Yes | Yes | No | Yes | No | Yes | Yes | No | Small molecule (< 2,000 a.m.u.) MM and QM tools to determine conformation, structure, property, spectra, reactivity, and selectivity. | Proprietary, free trial available | Wavefunction, Inc. |
| TeraChem | No | No | Yes | Yes | No | No | Yes | No | Yes | High performanceGPU-accelerated ab initiomolecular dynamics and TD/DFT software package for very large molecular or even nanoscale systems. Runs on NVIDIAGPUs and 64-bitLinux, has heavily optimized CUDA code. | Proprietary, trial licenses available | PetaChem LLC |
| TINKER | I | Yes | Yes | Yes | Yes | I | I | Yes | Yes | Software tools for molecular design-Tinker-OpenMM[10] Software tools for molecular design-Tinker-HP[11] | Proprietary, gratis | Washington University |
| Tremolo-X | I | No | Yes | Yes | No | No | No | No | No | Fast, parallel MD | Proprietary | Tremolo-X |
| UCSF Chimera | Yes | Yes | Yes | No | No | No | No | No | No | Visually appealing viewer, amino acid rotamers and other building, includes Antechamber and MMTK, Ambertools plugins in development. | Proprietary, free academic use | University of California |
| VEGA ZZ | Yes | Yes | Yes | I | Yes | No | No | Yes | Yes | 3D viewer, multiple file format support, 2D and 3D editor, surface calculation, conformational analysis, MOPAC and NAMD interfaces, MD trajectory analysis, molecular docking, virtual screening, database engine, parallel design, OpenCL acceleration, etc. | Proprietary, free academic use | VEGA ZZ website |
| VLifeMDS | Yes | Yes | Yes | No | Yes | No | I | No | Yes | Complete molecular modelling software, QSAR, combinatorial library generation, pharmacophore, cheminformatics, docking, etc. | Proprietary | Vlife Sciences Technologies |
| WHAT IF | Yes | Yes | I | I | I | No | No | No | No | Visualizer for MD, interface to GROMACS | Proprietary | WHAT IF |
| YASARA | Yes | Yes | Yes | Yes | No | No | Yes | No | Yes | Molecular graphics, modeling, simulation | Proprietary | YASARA.org |
See also[edit]
Notes and references[edit]
- ^M. J. Harvey, G. Giupponi and G. De Fabritiis (2009). 'ACEMD: Accelerating Biomolecular Dynamics in the Microsecond Time Scale'. Journal of Chemical Theory and Computation. 5 (6): 1632–1639. arXiv:0902.0827. doi:10.1021/ct9000685. PMID26609855.
- ^Johnston, MA, Fernández-Galván, I, Villà-Freixa, J (2005). 'Framework-based design of a new all-purpose molecular simulation application: the Adun simulator'. J. Comput. Chem. 26 (15): 1647–1659. doi:10.1002/jcc.20312. PMID16175583.CS1 maint: multiple names: authors list (link)
- ^Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995). 'A second generation force field for the simulation of proteins, nucleic acids, and organic molecules'. J. Am. Chem. Soc. 117 (19): 5179–5197. CiteSeerX10.1.1.323.4450. doi:10.1021/ja00124a002.
- ^Harrison ET, Weidner T, Castner DG, Interlandi G (2017). 'Predicting the orientation of protein G B1 on hydrophobic surfaces using Monte Carlo simulations'. Biointerphases. 12 (2): 02D401. doi:10.1116/1.4971381. PMC5148762. PMID27923271.
- ^Implicit Solvent - GromacsArchived July 29, 2014, at the Wayback Machine
- ^MAPS
- ^I.A. Solov'yov, A.V. Yakubovich, P.V. Nikolaev, I. Volkovets, A.V. Solov'yov (2012). 'MesoBioNano Explorer - A universal program for multiscale computer simulations of complex molecular structure and dynamics'. J. Comput. Chem. 33 (30): 2412–2439. doi:10.1002/jcc.23086. PMID22965786.CS1 maint: uses authors parameter (link)
- ^Macke T, Case DA (1998). 'Modeling unusual nucleic acid structures'. Molecular Modeling of Nucleic Acids: 379–393.
- ^A. Górecki; M. Szypowski; M. Długosz; J. Trylska (2009). 'RedMD - Reduced molecular dynamics package'. J. Comput. Chem. 30 (14): 2364–2373. doi:10.1002/jcc.21223. PMID19247989.
- ^M. Harger, D. Li, Z. Wang, K. Dalby, L. Lagardère, J.-P. Piquemal, J. Ponder, P. Ren (2017). 'Tinker-OpenMM: Absolute and relative alchemical free energies using AMOEBA on GPUs'. Journal of Computational Chemistry. 38 (23): 2047–2055. doi:10.1002/jcc.24853. PMC5539969. PMID28600826.CS1 maint: multiple names: authors list (link)
- ^L. Lagardère,L.-H. Jolly, F. Lipparini, F. Aviat, B. Stamm, Z. F. Jing, M. Harger, H. Torabifard, G. A. Cisneros, M. J. Schnieders, N. Gresh, Y. Maday, P. Y. Ren, J. W. Ponder, J.-P. Piquemal (2018). 'Tinker-HP: a massively parallel molecular dynamics package for multiscale simulations of large complex systems with advanced point dipole polarizable force fields'. Chemical Science. 9 (4): 956–972. doi:10.1039/C7SC04531J. PMC5909332. PMID29732110.CS1 maint: multiple names: authors list (link)